Image Rendering
In this document we'll cover how to view the contents of an image file.
Originally I began this document with the ide of just covering the
use of xpa to run ds9. When I began searching for ways to make images
with overplotted information, particularly markers having to do with
important spatial information (positions of star, N-E legend, plate
scale legend, etc...) I began to see that ds9 was limited. Actually, you
can do a lot of the operations with ds9, but if you want to do something
like make a more standard image format (like png) with ds9, and then
pretty it up with other graphics packages, you hit some fundamental
problems that (as of June2015) I have been unable to solve. Hence, I
added some sections on making image plots with python (specifically
matplotlib) and the seemingly pervasive ImageMagic package.
- Astronomical image files.
- Using ds9 and xpa.
- Using ImageMagic.
- Using python: matplotlib and other things.
- A mix of things for VIRUS.
- Nov2015 Commissioning Run.
Astronomical image files.
The basic image file format we'll see most in astronomy is a FITS files.
The ds9 display code is designed to handle FITS images, and the xpa code
is designed to communicate with the ds9 window. By communicate, I mean
your can execute ds9 commands from your script or program that allow you
to interact with your image. In this little intro section I show some
things we can do with a test image of NGC3379 (named n3379_B.fits). All
of this was done in my Test_Data directory: $tdata/T_runs/Image_Render/ex0_n3379.
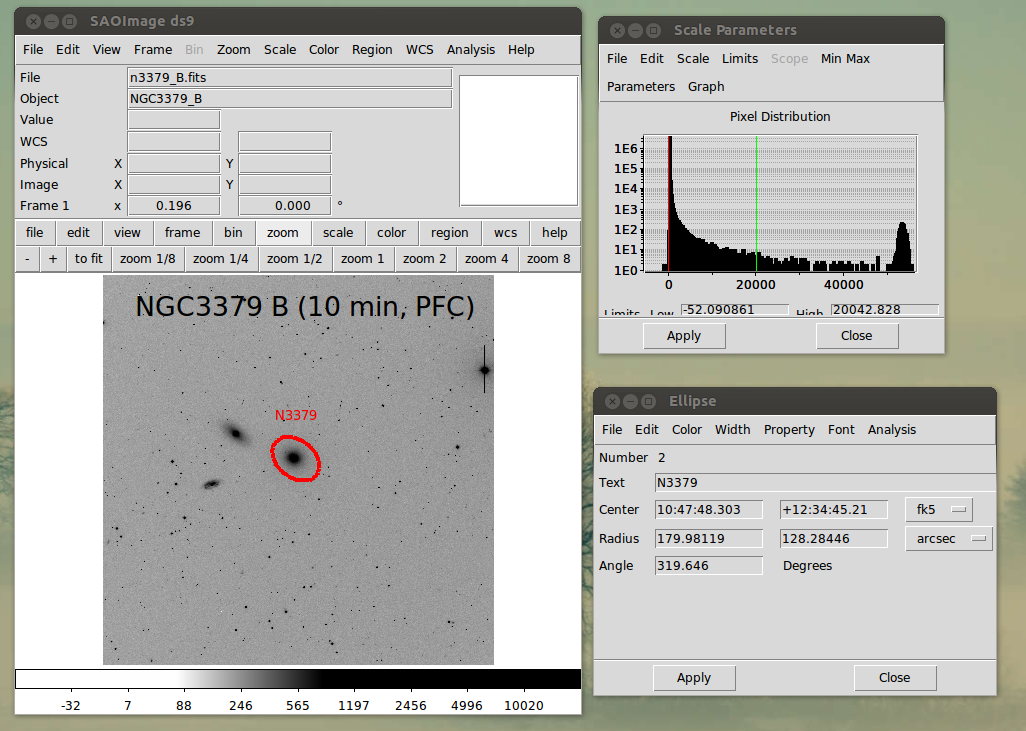 |
Here we use ds9 to view the image of NGC3379. I have
run ds9 in a manual mode here and I opened the Scale
window so that I could view the histogram of pixel
values and establish a better grey-scale image. I also
marked an ellipse region and I opened the region window
that describes that ellipse. Because the input image
has WCS parameters in the header, I can have the ellipse
(or any cursor properties) expressed in units like
RA, Dec, arcseconds, etc.... as well as he usual simple
pixel values. This is a major strength of ds9, it
can use a lot of the useful information present in
FITS headers. Finally, note that I made the html
text for this spiffy figure using:
build_htm_img ds9_manual.png ds9_manual.txt > my_html
|
I have written and collected a lot of tools that can operate
with FITS files. Below I show few snippets of code that take
our NGC 3379 image above, and create a png (Portable Network
Graphic) image file. I use xpa commands from within a script
to run ds9. The ds9 graphic above took about a minute to
execute. The one below took about a second. Moreover, I can
use this approach to compile a very complicated image construction
sequence, and use it over and over. Just bewause it is useful,
I show below how I used the getfits tool (from wcstools) to
cut out a small portion of the NGC3379 image for this plot. My
little script (called "doit") wants is hard-coded to use this
sub-image (called sub.fits) to create a png file (which I show you
below).
% getfits -o sub.fits n3379_B.fits 420-1020 740-1340
Row 1339 extracted
sub.fits
% ccdsize sub.fits
601 601
% doit 800 800
Enter any key AFTER ds9 window opens.
DS9 ds9 gs 7f000101:43417 sco
User z2 = 519.300
c_sub_800_800.png PNG 800x653 800x653+0+0 8-bit DirectClass 440KB 0.000u 0:00.000
Here are the guts of "doit":
#!/bin/bash
# Check command line arguments
if [ -z "$1" ]
then
printf "Usage: doit 500 700\n"
printf "arg1 - X size\n"
printf "arg2 - Y size\n"
exit
fi
ds9_open $1 $2
ds9_views_off
imlook sub.fits 30
xpaset -p ds9 saveimage c_sub_$1_$2.png
identify c_sub_$1_$2.png
Finally, just for completeness I will identify all three of the
bright galaxies in the rather lousy PFC image I am using here
in this exercise. Note that I use eye-ball settings of region
markers to create a regions file for the three galaxies and list
their Ra,Dec according to the image WCS. As a sanity check on the
getfits code (i.e. to insure that it is still preserving WCS properly)
I list the ds9 equatorial coordinates I derive from the sub.fits image.
NED Coordinates (J2000) Image WCS Sub_Image WCS
-------------------------- ------------------------- -------------------------
NGC3379 (E) 10h47m49.588s +12d34m53.85s 10:47:49.485 +12:34:54.07 10:47:49.485 +12:34:52.72
NGC3384 (S0) 10h48m16.886s +12d37m45.38s 10:48:16.628 +12:37:44.03 10:48:16.720 +12:37:45.39
NGC3389 (Sp) 10h48m27.910s +12d31m59.50s 10:48:28.029 +12:31:51.33 10:48:28.305 +12:31:54.05
For the full image: n3379_B.fits
X_image Y_image
NGC3379 (E) 974.0 1055.5
NGC3384 (S0) 680.0 1180.5
NGC3389 (Sp) 557.0 925.5
Finally, for the full image, I can draw a large circle
that goes through the centers of {NGC3379,84,89} with:
CENTER : Ra,Dec=10:48:08.297,+12:33:20.00 X,Y=(771.0,986.5)
RADIUS = 292.7 arcsec , 216.1 pixels
I list the generic galaxy types to make them easily identifiable. The
big circular E galaxy (on the RIGHT) is N3379. The elliptical-shaped
S0 galaxy (at the TOP) is N3384, and the late-type spiral (lower-LEFT)
is N3389. It is interesting to note that these 3 fairly
bright, distinctly different (morphologic and spectral) galaxies would
fill a field about 10' x 10' on the sky: a size that would be nicely
accomodated by the VIRUS spectrograph on HET.
 |
|
Here is the image of NGC3379 using the doit script
described above. As we'll see later in this document, I can
use xpa to make all sorts of adjustemnts, and then create a
more general image file (like png, tiff, jeg,...). However,
one problem that I have encountered is evident above. The
user specifies the size of the entire ds9 gui (800x800 above), but
the image is displayed in the viewing windo area, and I have been
unable to learn how one can query or set this size. The problem
arises because when we dump and image file (using saveimage in
ds9), the ENTIRE viewing window is dumped, including all of the
surrounding white-space! As we'll see later in this document, this
complicates the process of using other method (like a python
script) to over-write things on our image file. Such is life.
|
Finally, I can take a file like the png image we generated above
and spiff it up using the python matplotlib package:
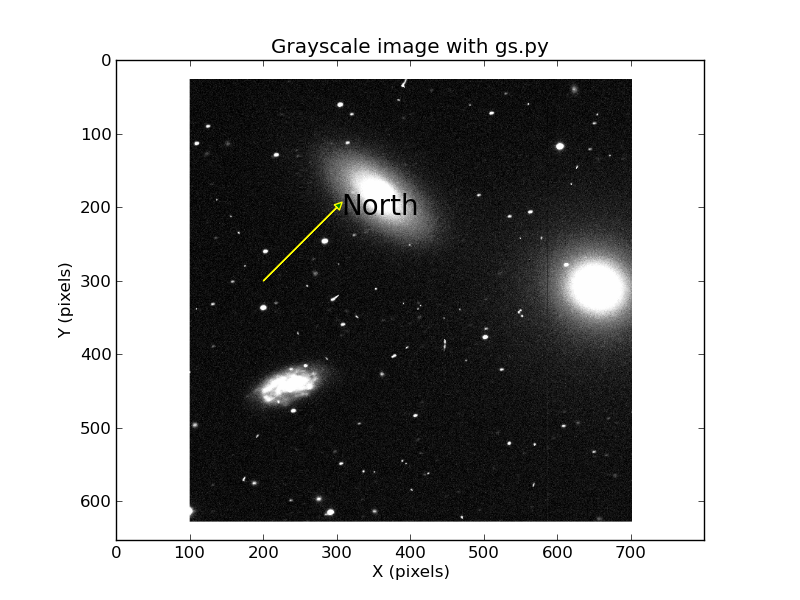 |
Here is the type of image I can generate using a bone-head simple
python script that utilized the matplotlib package. I've taken
the png file from the above exercise and run the command:
% gs.py c_sub_800_800.png
Of course, the overplotted markers and text are nonsense and
used here only as a demonstration. An important word of WARNING:
if you are a good ds9-er, you'll know that normally the image
will be displayed with +X pixels going to the right, and +Y
pixels going to towards the top of the display. This is
the way our pixel coordinates will be read in our very top
NGC3379 graphic above. Notice that the configuration
of stars and galaxies around NGC3379 (the round galaxy) is
the same in all of our images. However, the Y-axis pixels
are labeled above in our python version with +Y values
increasing towrads the bottom. We'll have to be
mindful of such things when creating things like finding charts, etc..
with packages like matplotlib. Some information on how gs.py works in
this imshow tutorial.
|
As with all things python, the documentation for matplotlib is terrible.
However, since the latest ds9+xpa online documentation is comparably
poor, we can jump into the python graphics abyss with enthusiastic
abandon.
Using ds9 and xpa.
This is a broad topic, but a good place to start is
with some of my own xpa
and ds9 notes. The ds9 rendering approach is good when you
will want to interact with the image you have overplotted. For
instance, you might have overplotted the fiber positions in a VIRUS
IFU, but you want to zoom into a small area to view some fine detail.
I have begun a set of higher level tools that use xpa+ds9 to
do a variety of rendering jobs for VIRUS. Sample data and
early README files are located in the TDATA/Image_Render/ex0_n3379/S
directory. I'll just summarize the appraoch here:
- I have a number coded that create the basic types of plot information:
the IHMP outline (ihmp.sh), the VIRUS IFU pattern (vir_ifu_pat.sh), the
outline of the HET guide probe annulus (het_gp_pickle.sh). The basic
XY data sets made with these tools are in units of arcseconds, and
have an origin at 0,0. I generally refere to these as APERTURE files.
- The job of preparing aperture APERTURE file for a particular image
involves a scale change, rotation, and translation. These produce
files with X,Y in pixel units, and they are typified by codes like
cbf_trans.sh (closed boundary files, for the IHMP), lsegs_trans.sh
(line segments, e.g. for IFU bounds or guide probe annuli),
text_trans.sh (for marking text, e.g. for labeling IFU names).
- The actual preparation of the ds9 region files is done with
python scripts (.../codes/python/ds9 ) like: xyr_circles_ds9.py,
lines_file_ds9.py, lsegs_file_ds9.py, text_file_ds9.py. Some of
these routines take parameters on the command line and produce
a region string, some take the name of a fle and produce strings
for multiple regions.
Some early examples using these tools
are shown here.
IMPORTANT VIRUS TOOL NOTES: I had a
lot of notes on routines I developed to overplot VIRUS and HET features
on DSS (and other ) images. These notes, and support run scripts are all in:
Test_Data_for_Codes/T_runs/Image_Render/ex0_n3379/S
Because I always seem to loose these things (I spent an hour one night trying to
remember how to plot guide probe annuli, and then I found notes about the
routine het_gp_pickle.sh) I have now assembled all of these notes into
a single big file of notes about
image rendering routines using (mostly) ds9+xpa.
Using ImageMagic.
The ImageMagic routines (mostly convert) can be used to perform a
many simple (and not so simple) image-processing tasks. I would never
trust this stuff to process images for science purposes, but it sure is handy
sometimes to get things into cartoon-like state for building figures.
Moreover, it seems to come intsalled with almost every linux distribution
you can think of. Some discussions and
Here are detailed ImageMagic examples
are here. The bottom line to date is that I DO NOT UNDERSTAND much
of the image processing tasks employed in the ImageMagic convert task. I
spent the better part of an evening and morning reading web material, and
it is clear that some important concepts elude me. I think the primary
confusion has to do with the fact that the PNG and other standard rendering
files assume 8-bit (2^8 ---> 0 to 256) pixel ranges. Moreover, a lot of
the special transform features ("-fx") assume ranges of 0 to 1. What seems
to be missing inthe online stuff (to me at least!) is any kind of reasonable
explanations of how you take images with larger ranges (i.e. FITS images
with BITPIX=-32 floating point pixels) and prep them to the point of using
convert on them. So, I move on for now and investigate other routes to
rendering astronomical images with a useful dynamic range.
Using python: matplotlib and other things.
Originally I envisioned a document here that just described how
I "make an image" from something in a FITS file. That is clearly
the overall goal, but I have found after my experience with ImageMagic,
that some other capabilities of python may be put to very good use
now. Specifically, viewing pixel histograms (standard and cummulative)
will help me to understand the basics of image display. I can use the
gs.py code described above in my introduction, but getting to the
useful PNG file that this code needs is a bigger subject. In the
following link I
discuss image rendering using python.
The real work is in establishing the transfer function that gets my
FITS pixels into the 8-bit (0 to 256) range (for instance) that makes for
a good PNG image.
A mix of things for VIRUS.
A variety of tool sets have been combined to create a
general-purpose rendering tool for VIRUS. The properties
of this tool are
described here.
Nov2015 Commissioning Run.
I describe software tools
used in the Nov2015 LRS2 commissioning run. This is meant
to record some of the methodology used during early commissioning
tests and does not imply anything about future observing procedures.
Back to SCO CODES page


