- Initial Observing Notes
- A quick survey of the acm image sets
- A master set of acm targets.
- Measuring the acm image sets.
- Measuring the gc1 image sets.
- Measuring the gc2 image sets.
- Plate scales are measured and plotted.
Initial Observing Notes
On 20190724 we (SCO/NM) made observations designed for deriving the gc1,gc2 plate scale. We
took sets of acm,gc1,gc2 images with guiding done on acm. We moved the acm fiducial to
to perform the telescope offsets. We used about 7 offsets, with the last two done with
no "pid loop".
Original notes made by SCO:
==================================================================================
20190724
A: SCO TO: NM
===========================================================
ENG gc offset test to derive plate scale
Moon is about 20deg above horizon, and 60% illum
target_setup 54 E -cat gp -ifu 000
This field: 54 GALl110bm5 23:18:39.51 +55:31:29.69 2000 34.3 15.00 06:51 02:11 07:49 01:55
To make acm offsets:
syscmd -T 'ACQ_offset_fiducial ( dx_asec=5,dy_asec=0,compensate="false")'
Go to target at 06:55 UT
Note DIMM seeing is jumping to 2" just as we get going on this test!
Position START
Starting set of 10 at 07:02:22 UT (stars centered in gc1,gc2)
Stop taking images at at 07:04:32
Position A
At 7:05
syscmd -T 'ACQ_offset_fiducial ( dx_asec=5,dy_asec=0,compensate="false")'
*** star moves straight down on gc2
wait for focus and guider stability
Start taking images at 07:08:06 UT
stop 07:10:21
Position B
At 7:10:52
syscmd -T 'ACQ_offset_fiducial ( dx_asec=-10,dy_asec=0,compensate="false")'
star moves over, but stys well within gc1,gc2
wait for focus and guider stability
Start taking images at 07:14:24 UT
stop 07:16:53
Position C
At 7:17:10
syscmd -T 'ACQ_offset_fiducial ( dx_asec=+5,dy_asec=-6,compensate="false")'
wait for focus and guider stability
Start taking images at 07:20:30 UT
stop at 07:22:47
Position D
At 07:23:20
syscmd -T 'ACQ_offset_fiducial ( dx_asec=0,dy_asec=+12,compensate="false")'
syscmd -T 'ACQ_offset_fiducial ( dx_asec=0,dy_asec=+2,compensate="false")'
wait for focus and guider stability
Start taking images at 07:30:15 UT
stop at 07:32:29
Position E
At 07:33:25
syscmd -T 'ACQ_offset_fiducial ( dx_asec=0,dy_asec=-14,compensate="false")'
wait for focus and guider stability
Start taking images at 07:37:00 UT
stop at 07:39:20
DIMM is hopping up to 2" again
With pid set to ZERO
Position F (same as E, but pid is off)
wait for focus and guider stability
Start taking images at 07:41:00 UT
stop at 07:43:33
Position G
At 07::43:54 (pid still off)
syscmd -T 'ACQ_offset_fiducial ( dx_asec=0,dy_asec=+14,compensate="false")'
Start taking images at 07:45:55 UT
stop 07:48:30
Done with test
===========================================================
**** Thu Jul 25 14:00:54 CDT 2019
I have moved the 20190724 acm,gc1,gc2 full data sets to:
/media/sco/DataDisk1/sco/AD/HET_work/acm_nights/20190724
(I have also move this to the steveo laptop)
The galpoint field (gp) I used for this is at: 23:18:39.51 +55:31:29.69
Recall that "galpoint" fileds are a list of low galactic latitude fields that
are listed in the htopx "gp" catalog. They are fields we use when we want a
hight number of stars in the field.
To get support data:
% dssbase 23:18:39.51 +55:31:29.69 6.0 19.5 N
For a non-internet reduction I can use: /home/sco/DSS_Base/acm_fields/Store_DSS
Here is a summary of the positions and their time intervals.
Position Set Recorded UT Interval
------------ -----------------------
START 07:02:22.0 07:04:32.0
A 07:08:06.0 07:10:21.0
B 07:14:24.0 07:16:53.0
C 07:20:30.0 07:22:47.0
D 07:30:15.0 07:32:29.0
E 07:37:00.0 07:39:20.0
F 07:41:00.0 07:43:33.0
G 07:45:55.0 07:48:30.0
==================================================================================
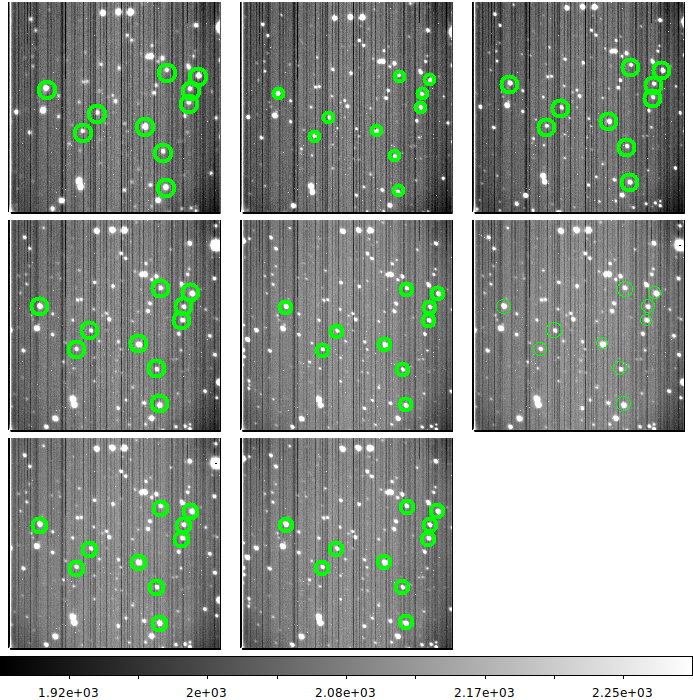
A quick survey of the acm images for the night
You can read about how how I reviewed the images in each position/time interval
in a discussionn of processing acm images.
Here I want to review each set of acm images and delete images that show large
position departures. I then use the final image listing to establish a new
time interval for each position. The basic recipe is:
I perform this in: /home/sco/GC_Plate_Scales/20190724
To get the initial catalog with image types:
acm_table_markII N
select_pas_images_by_time ACMDAT.images ACMDAT 07:02:22 07:04:32 STARTacm N
bigds9 STARTacm.images 1 20
mkdir STARTacm
mv ds9_list_load.nomark STARTacm/list.0
select_pas_images_by_time ACMDAT.images ACMDAT 07:08:06.0 07:10:21.0 Aacm N
bigds9 Aacm.images 1 20
mkdir Aacm
mv ds9_list_load.nomark Aacm/list.0
select_pas_images_by_time ACMDAT.images ACMDAT 07:14:24.0 07:16:53.0 Bacm N
bigds9 Bacm.images 1 20
mkdir Bacm
mv ds9_list_load.nomark Bacm/list.0
select_pas_images_by_time ACMDAT.images ACMDAT 07:20:30.0 07:22:47.0 Cacm N
bigds9 Cacm.images 1 20
mkdir Cacm
mv ds9_list_load.nomark Cacm/list.0
select_pas_images_by_time ACMDAT.images ACMDAT 07:30:15.0 07:32:29.0 Dacm N
bigds9 Dacm.images 1 20
mkdir Dacm
mv ds9_list_load.nomark Dacm/list.0
select_pas_images_by_time ACMDAT.images ACMDAT 07:37:00.0 07:39:20.0 Eacm N
bigds9 Eacm.images 1 20
mkdir Eacm
mv ds9_list_load.nomark Eacm/list.0
select_pas_images_by_time ACMDAT.images ACMDAT 07:41:00.0 07:43:33.0 Facm N
bigds9 Facm.images 1 20
mkdir Facm
mv ds9_list_load.nomark Facm/list.0
select_pas_images_by_time ACMDAT.images ACMDAT 07:45:55.0 07:48:30.0 Gacm N
bigds9 Gacm.images 1 20
mkdir Gacm
mv ds9_list_load.nomark Gacm/list.0
I reject (mark) images with large motions indicated and then rename the list of
unamrked image to an appropriate name. I create a subdirectory with this image file
and record the final range of UT times. These final UT time intervals will be used
to gather the corresponding gc1 and gc2 image sets.
Here is a summary of the refined time intervals for each position.
Position Set Refined UT Interval
------------ -----------------------
START 07:02:39.7 07:04:18.3
A 07:08:31.0 07:10:09.6
B 07:14:50.5 07:16:43.1
C 07:20:56.0 07:22:34.7
D 07:30:32.0 07:32:24.7
E 07:37:19.4 07:39:12.1
F 07:41:18.4 07:43:25.0
G 07:46:13.5 07:48:20.3
The above procedure is practical if we have only one or two position sets. In this
case we have 8 sets and performing all of these steps manually is laborous.
A BETTER and SAFE way to do this:
A better way to gather and visually review the images in each position set
is to use pas_time_window_sets.
Basically, I build one input file (along with the usual BaseDir and Date files) that
specieis each position name and time inetrval. For each set the names of images are
collected and run through bigds9, allowing
the user to reject bad images. Note that pas_time_window_sets is not specific to
acm images. Any type of PAS image can be treated. We'll use it to collect the
gc1 and gc2 images fro the corresponding positions. The code will form subdirectory
for each set where the cleaned image lists are stored. Finally, in each set
subdirectory the images are stacked to give a single image that represents the mean
field. Belwo I show how I processed the 8 positions for the acm images from 20190724.
% pwd
/home/sco/GC_Plate_Scales/20190724_red2
% ls
BaseDir Date S/ Time.Sets
% cat BaseDir
/media/sco/DataDisk1/sco/AD/HET_work/acm_nights
% cat Date
20190724
% cat Time.Sets
START 07:02:22.0 07:04:32.0
A 07:08:06.0 07:10:21.0
B 07:14:24.0 07:16:53.0
C 07:20:30.0 07:22:47.0
D 07:30:15.0 07:32:29.0
E 07:37:00.0 07:39:20.0
F 07:41:00.0 07:43:33.0
G 07:45:55.0 07:48:30.0
% pas_time_window_sets acm Time.Sets N
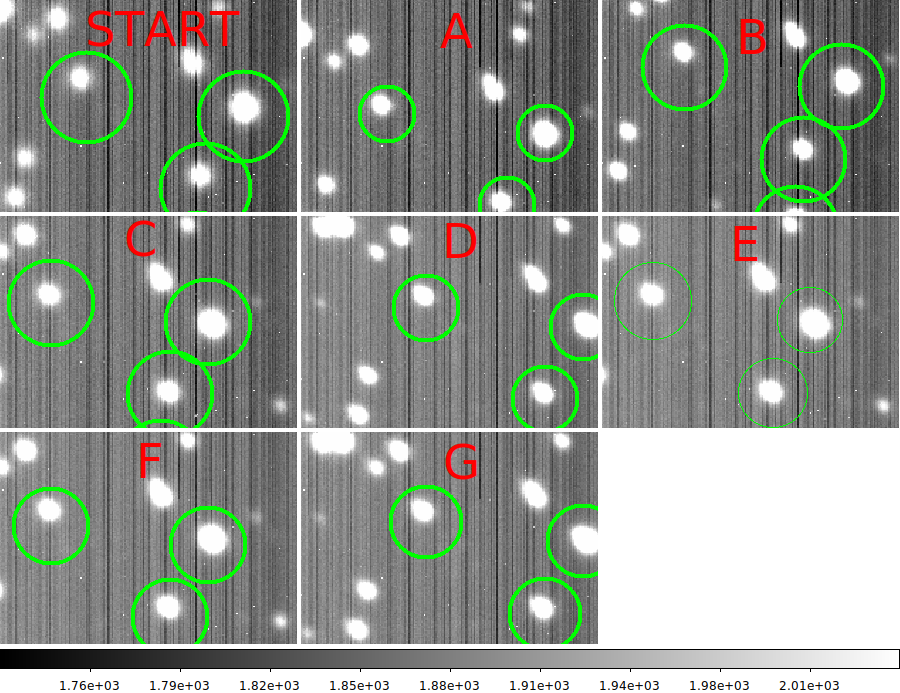
Note that as a sanity check I usually some in on a bright star and blink
the zoomed in image. When the set is completed, and I get the message like
"Set with name = A has been completed.", I manually record the X,Y position
of this star is eacj image set. I often find these rough positions useful in
verifying more precise offset calculations downstream in the reduction. I usually
just copy the Time.Sets file and fill in these XY values as I process each set.
START 07:02:22.0 07:04:32.0 499.2 316.1
A 07:08:06.0 07:10:21.0 500.6 298.0
B 07:14:24.0 07:16:53.0 500.8 335.6
C 07:20:30.0 07:22:47.0 478.6 315.0
D 07:30:15.0 07:32:29.0 529.4 315.6
E 07:37:00.0 07:39:20.0 475.8 316.9
F 07:41:00.0 07:43:33.0 476.9 317.0
G 07:45:55.0 07:48:30.0 530.0 315.8
NOTE: Positions E and F appear to be the same! So, I could disregard F.
At the end of this run, my directory looks like:
% ls
Aacm/ acmALL.params acmALL.table BaseDir Dacm/ ds9_open.Size Facm/ list.IMAGES STARTacm/
acmALL.images acmALL.parlab Bacm/ Cacm/ Date Eacm/ Gacm/ S/ Time.Sets
A master set of acm targets.
Typically I will also build a mosiac of the stacked fields and use the first image to
set up a set of acm stars that I will measure in each image set. For the subsequent sets
I use the ds9_region_xyshift.sh script
to apply XY shifts that will transform the XY coordinates of the original targets to
the coordinate system of each position set.
% pwd
/home/sco/GC_Plate_Scales/20190724_red2/figs_acm
% ls -1 ../*acm/*.fits >list.acm
**** I might edit list.acm *****
% bigds9 list.acm 1 10
***
I blink the images to locate stars that do not leave the field
or get too close to the image edge to allow centroid calculation.
I save a set of regions for a good image, in this example I save: Eacm.reg
I note the FIRST object I marked in this chart. This is the object I will
identify in all subsequent images for which I'll to build a shifted region file.
Then I create shifted versions of Eacm.reg for each image:
% ds9_region_xyshift.sh ./Eacm.reg ../STARTacm/STARTacm.fits N
% ds9_region_xyshift.sh ./Eacm.reg ../Aacm/Aacm.fits N
% ds9_region_xyshift.sh ./Eacm.reg ../Bacm/Bacm.fits N
% ds9_region_xyshift.sh ./Eacm.reg ../Cacm/Cacm.fits N
% ds9_region_xyshift.sh ./Eacm.reg ../Dacm/Dacm.fits N
% ds9_region_xyshift.sh ./Eacm.reg ../Eacm/Eacm.fits N
% ds9_region_xyshift.sh ./Eacm.reg ../Facm/Facm.fits N
% ds9_region_xyshift.sh ./Eacm.reg ../Gacm/Gacm.fits N
At the end of this process I have the region files for each image:
Aacm.fits Bacm.reg Dacm.reg Eacm.fits Facm.reg list.acm STARTacm.reg
Aacm.reg Cacm.reg ds9_open.Size Eacm.reg Gacm.reg S/
Now I can rerun bigds9 and then manually load each region file in the
appropriate frame. I usually copy each region file to its appropriate
subdirectory (with a script).
% bigds9 list.acm 1 10
The two figures below illustrate this mosaic process with bigds9.
The position subdirectories gathered above now contain a list of images
(list.0), a mean stacked image of the field, and an appropriately shifted
region file. In the case of this reduction, each field will have the same
10 stars measured in each image of each position set. For each subdirectory
I would run command sequence like that below:
To measure position offsets
To compute the possible sets of offsets:
cd /home/sco/GC_Plate_Scales/20190724_red2
table_XY_offsets.sh ./Aacm/XYmean ./Bacm/XYmean xmean ymean xme yme N
pas_XY_offsets.sh ./Aacm/list.0 ./Bacm/list.0 1.0 N
An easy script run: ./S/ORUN Aacm Bacm
./S/ORUN STARTacm Eacm
./S/ORUN Cacm Dacm
./S/ORUN Gacm Facm
Here I collect the acm position set offsets
SCO centroid PAS centroids Position Pair
mean median m.e. Nstar mean median m.e. Nims
--------------------------------------- ---------------------------------------
35.719 35.959 0.293 10 37.156 37.256 0.519 8 A-B
22.979 22.745 0.352 10 21.989 22.293 0.803 8 START-E
52.101 52.203 0.112 10 52.159 52.421 0.454 8 C-D
51.679 51.715 0.068 10 51.658 51.751 0.494 10 G-F
Measuring the gc1 image sets.
Because the gc images normally posess only a single bright star, the
mesaurement of the images is less complicated than the acm sets.
*** Gather the position set subdirectories ********
There are N=3423 gc1 images
% pwd
/home/sco/GC_Plate_Scales/20190724_red2
% ls
BaseDir Date S/ Time.Sets
% cat BaseDir
/media/sco/DataDisk1/sco/AD/HET_work/acm_nights
% cat Date
20190724
% cat Time.Sets_refined
START 07:02:39.7 07:04:18.3
A 07:08:31.0 07:10:09.6
B 07:14:50.5 07:16:43.1
C 07:20:56.0 07:22:34.7
D 07:30:32.0 07:32:24.7
E 07:37:19.4 07:39:12.1
F 07:41:18.4 07:43:25.0
G 07:46:13.5 07:48:20.3
% pas_time_window_sets gc1 Time.Sets_refined N
% pwd
/home/sco/GC_Plate_Scales/20190724_red2/figs_gc1
% ls -1 ../*gc1/*.fits >list.gc1
**** I might edit list.acm *****
% bigds9 list.gc1 1 10
*** In each subdirectory for gc1
% ds9_imstats_fitslist list.0 FixedRegions N
To compute the possible sets of offsets:
cd /home/sco/GC_Plate_Scales/20190724_red2
table_XY_offsets.sh ./Agc1/XYmean ./Bgc1/XYmean xmean ymean xme yme N
pas_XY_offsets.sh ./Agc1/list.0 ./Bgc1/list.0 1.0 N
An easy script run: ./S/ORUN Agc1 Bgc1
./S/ORUN STARTgc1 Egc1
./S/ORUN Cgc1 Dgc1
./S/ORUN Ggc1 Fgc1
Here I collect the acm position set offsets
SCO centroid PAS centroids Position Pair
mean median m.e. Nstar mean median m.e. Nims
--------------------------------------- ---------------------------------------
51.748 51.748 0.403 1 51.296 51.381 0.532 20 A-B
32.049 32.049 0.501 1 32.001 31.901 0.660 20 START-E
71.346 71.346 0.421 1 71.235 71.287 0.466 19 C-D
71.227 71.227 0.417 1 70.705 70.868 0.521 24 G-F
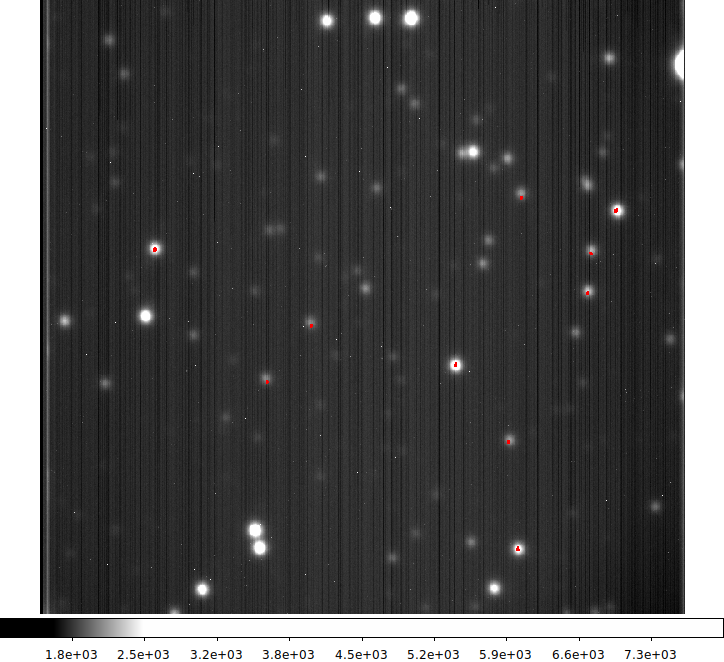
 |
|
The mean (stacked) gc1 images. The position set is indicated by the large red letters.
|
Measuring the gc2 image sets.
Here I present the same procedure for gc2.
*** Gather the position set subdirectories ********
There are N=3821 gc2 images
% pwd
/home/sco/GC_Plate_Scales/20190724_red2
% ls
BaseDir Date S/ Time.Sets
% cat BaseDir
/media/sco/DataDisk1/sco/AD/HET_work/acm_nights
% cat Date
20190724
% cat Time.Sets_refined
START 07:02:39.7 07:04:18.3
A 07:08:31.0 07:10:09.6
B 07:14:50.5 07:16:43.1
C 07:20:56.0 07:22:34.7
D 07:30:32.0 07:32:24.7
E 07:37:19.4 07:39:12.1
F 07:41:18.4 07:43:25.0
G 07:46:13.5 07:48:20.3
% pas_time_window_sets gc2 Time.Sets_refined N
% pwd
/home/sco/GC_Plate_Scales/20190724_red2/figs_gc2
% ls -1 ../*gc2/*.fits >list.gc2
**** I might edit list.acm *****
% bigds9 list.gc2 1 10
*** In each subdirectory for gc1
% ds9_imstats_fitslist list.0 FixedRegions N
To compute the possible sets of offsets:
cd /home/sco/GC_Plate_Scales/20190724_red2
table_XY_offsets.sh ./Agc1/XYmean ./Bgc1/XYmean xmean ymean xme yme N
pas_XY_offsets.sh ./Agc2/list.0 ./Bgc2/list.0 1.0 N
An easy script run: ./S/ORUN Agc2 Bgc2
./S/ORUN STARTgc2 Egc2
./S/ORUN Cgc2 Dgc2
./S/ORUN Ggc2 Fgc2
Here I collect the acm position set offsets
SCO centroid PAS centroids Position Pair
mean median m.e. Nstar mean median m.e. Nims
--------------------------------------- ---------------------------------------
51.715 51.715 0.383 1 51.617 51.526 0.477 20 A-B
33.071 33.071 0.498 1 32.604 32.629 0.627 20 START-E
71.238 71.238 0.385 1 71.347 71.620 0.457 20 C-D
71.079 71.079 0.398 1 71.171 71.451 0.520 24 G-F
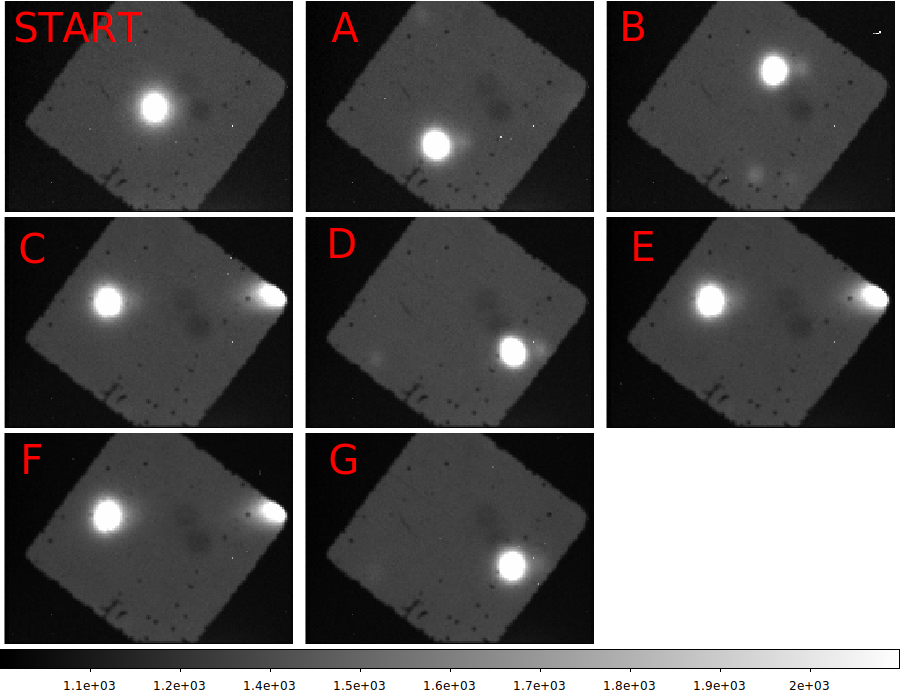
 |
|
The mean (stacked) gc2 images. The position set is indicated by the large red letters.
|
Plate scales are measured and plotted.
The plate scale values for the offsets are computed.
I collect offsets in
% pwd
/home/sco/GC_Plate_Scales/20190724_red2/PS_extimates
Build a file names 20190724.dat:
% cat 20190724.dat
A-B
35.719 35.959 0.293 10 37.156 37.256 0.519 8 A-B
51.748 51.748 0.403 1 51.296 51.381 0.532 20 A-B
51.715 51.715 0.383 1 51.617 51.526 0.477 20 A-B
START-E
22.979 22.745 0.352 10 21.989 22.293 0.803 8 START-E
32.049 32.049 0.501 1 32.001 31.901 0.660 20 START-E
33.071 33.071 0.498 1 32.604 32.629 0.627 20 START-E
C-D
52.101 52.203 0.112 10 52.159 52.421 0.454 8 C-D
71.346 71.346 0.421 1 71.235 71.287 0.466 19 C-D
71.238 71.238 0.385 1 71.347 71.620 0.457 20 C-D
G-F
51.679 51.715 0.068 10 51.658 51.751 0.494 10 G-F
71.227 71.227 0.417 1 70.705 70.868 0.521 24 G-F
71.079 71.079 0.398 1 71.171 71.451 0.520 24 G-F
Next we compute the plate scale
% gcps.sh 20190724.dat N
To make a plot of the estimates (for example):
% Generic_Points N
% xyplotter_auto gc1_plate_scales q q 1 N
(edit Axws.1 List.1)
% xyplotter List.1 Axes.1 N
I add the plot data to my older 3 set analysis in: /home/sco/GC_Plate_Scales/4sets
xyplotter xyplotter List.1 Axes.1 N
xyplotter xyplotter List.2 Axes.2 N
Here are the stats for ALL data from stable nights:
GC1:
% cat gc1_stable.dat
0.1973
0.1989
0.1989
0.1870
0.1886
0.1942
0.1945
0.1978
0.1981
0.1966
0.1980
% calstats.py -v gc1_stable.dat
Simple stats for numbers in: gc1_stable.dat
Mean = 0.19545
Median = 0.19730
Standard deviation = 0.00391
Minimum = 0.18700
Maximum = 0.19890
Number of values = 11
Mean error of then mean = 0.00124
GC2:
% cat gc2_stable.dat
0.1948
0.1971
0.1975
0.1871
0.1875
0.1882
0.1909
0.1981
0.1978
0.1970
0.1967
% calstats.py -v gc2_stable.dat
Simple stats for numbers in: gc2_stable.dat
Mean = 0.19388
Median = 0.19670
Standard deviation = 0.00430
Minimum = 0.18710
Maximum = 0.19810
Number of values = 11
Mean error of then mean = 0.00136
Finally, I derive weighted mean stats for only the 3 stable nights:
% pwd
/home/sco/GC_Plate_Scales/4sets/Only_Stable_Nights
% table_stats.sh PS_gc1 ps pserr N
mean,median,sigma,m.e.,n:
0.195445 0.197300 0.004103 0.001237 11
Weighted estimates (mean,sigma,m.e):
0.196391 0.003855 0.001162
% table_stats.sh PS_gc2 ps pserr N
mean,median,sigma,m.e.,n:
0.193882 0.196700 0.004506 0.001358 11
Weighted estimates (mean,sigma,m.e):
0.195371 0.004015 0.001211
**** Here are the final data file I used for the above stats
% cat PS_gc1.table
PS values from stable data nights
Pair cam meth Sep ps m.e. date
# data
AB gc1 sco 12.1 0.1973 0.0015 20190711
AB gc1 sco 10.2 0.1989 0.0012 20190720
AB gc1 pas 10.2 0.1989 0.0013 20190720
AB gc1 sco 9.7 0.1870 0.0015 20190724
AB gc1 pas 9.7 0.1886 0.0020 20190724
STE gc1 sco 6.2 0.1942 0.0030 20190724
STE gc1 pas 6.2 0.1945 0.0040 20190724
CD gc1 sco 14.1 0.1978 0.0012 20190724
CD gc1 pas 14.1 0.1981 0.0013 20190724
GF gc1 sco 14.0 0.1966 0.0012 20190724
GF gc1 pas 14.0 0.1980 0.0015 20190724
% cat PS_gc2.table
Stable night gc2 estimates
Pair cam meth Sep ps m.e.
# data
AB gc2 sco 12.1 0.1948 0.0016 20190711
AB gc2 sco 10.2 0.1971 0.0011 20190720
AB gc2 pas 10.2 0.1975 0.0013 20190720
AB gc2 sco 9.7 0.1871 0.0014 20190724
AB gc2 pas 9.7 0.1875 0.0017 20190724
STE gc2 sco 6.2 0.1882 0.0028 20190724
STE gc2 pas 6.2 0.1909 0.0037 20190724
CD gc2 sco 14.1 0.1981 0.0011 20190724
CD gc2 pas 14.1 0.1978 0.0013 20190724
GF gc2 sco 14.0 0.1970 0.0011 20190724
GF gc2 pas 14.0 0.1967 0.0014 20190724
The parlab files:
% cat PS_gc2.parlab
posp Name of Position Pair
cam GC camera name
meth Method (Sco centroids or PAS)
sep Star Separation in arcseconds
ps plate scale (arcsec/pixel)
pserr mean error of plate scale (arcsec/pixel)
date Date of the Observations (UT)
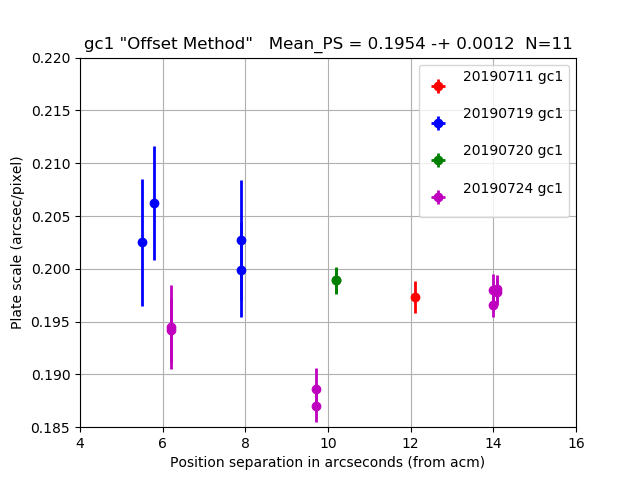
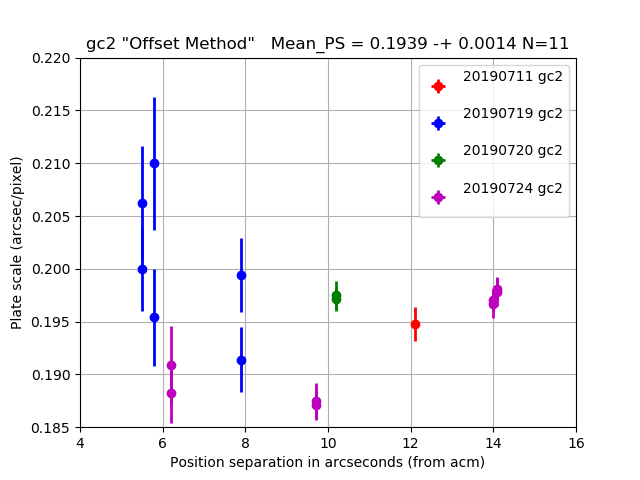
The 11 "stable night" values are used to compute the mesn values in the titles of
the plots below.
 |
|
The gc1 plate scale from 4 nights of offset tests.
|
 |
|
The gc2 plate scale from 4 nights of offset tests.
|
Back to calling page