Exercise 2: A WCS exercise built with usno_identify.
Here I use usno_identify to build the test file. Here I create the type
of file I refer to as a *.xyrd file. It contains the same info as the
wcs_setup.out file, but the data for each source are on a single line.
I want to use my n4625 acam stack and create a *.xyrd file to use in testing wcs_markII.
I do this when the internet is down, so I use my stored dss image of n4625:
% ls $critfils/tdat
ds9_n4625.png dss_n4625.fits hetacm_n4625.fits README.Test_n4625 wcs1/
% cp $critfils/tdat/hetacm_n4625.fits .
% cp $critfils/tdat/dss_n4625.fits dss.fits
Next, because I know this image has an old, very wrong, tcs header, I remove all wcs info
from the header of this image:
% remove_wcs hetacm_n4625.fits
After doing this you should view the image with ds9 and confirm that only X,Y value
are displayed. No Ra,Dec values appear because the WCS information, if present, was
removed from the header.
Then I run some quicky tests:
% wcs_present_in_header hetacm_n4625.fits
N
My wcs info is gone, so I'll need some basic info for when I run usno_identify. I need
RA,DEC and a field size. I use rc3gal to get the position for n4625, and I know a good
field size for the acm is 4.0 arcminutes.
Results of the RC3 search:
Name_RC3 RA_2000DEC_2000 Type Input_name
NGC 4625 12:41:53.00 +41:16:27.0 .SXT9P. n4625
dssget 190.470825 41.274166 10.0 dss poss1_red
So I'll be entering:
12:41:53.00 +41:16:27.0 4.0
To run:
% usno_identify hetacm_n4625.fits 20.0 blue 16
* Because there is no WCS header info I must manually enter info about
where the image is on the skay and the size of the image. I use:
RAsex_,DECsex, Rad_ArcMin = 12:41:53.00 +41:16:27.0 4.0
NOTE: Originally I used my stored dss image, but this turned out to be just
a little to small for my acm stacked image. So, I ran this exercise when the
internet wa available and I pulled a large dss field.
I had to flip the image orientation (I flipped the Y axis) to get an
alignment that allowed me to match stars.
*** I made the file: usno_identify.final_list
% cat usno_identify.final_list
565.16 497.97 12:41:43.3608 +41:15:47.3099 10
433.35 455.45 12:41:46.4316 +41:16:00.3287 13
665.79 410.09 12:41:40.7364 +41:16:05.7806 8
704.05 255.61 12:41:39.4332 +41:16:48.5037 7
204.89 119.84 12:41:51.0648 +41:17:37.1182 18
640.43 740.80 12:41:42.1620 +41:14:36.9974 9
333.69 73.92 12:41:47.7852 +41:17:46.0446 15
--------------------------------------------------------------------
NOTE: For an *.xyrd file:
% make_xyrdfile hetacm_n4625.fits usno_identify.final_list
*** May embed this into usno_identify
Format I need:
241.040 1115.340 346.367767334 12.421183586 194 23:05:28.26 +12:25:16.3
703.170 258.290 190.414305000 41.280139917 1 12:41:39.4332 +41:16:48.5037
This makes the file = hetacm_n4625.xyrd
% make_xyrdfile hetacm_n4625.fits usno_identify.final_list
% cat hetacm_n4625.xyrd
# Sources collected with make_xyrdfile
# data
565.160 497.970 190.430670000 41.263141638 10 12:41:43.3608 +41:15:47.3099
433.350 455.450 190.443465000 41.266757972 13 12:41:46.4316 +41:16:00.3287
665.790 410.090 190.419735000 41.268272389 8 12:41:40.7364 +41:16:05.7806
704.050 255.610 190.414305000 41.280139917 7 12:41:39.4332 +41:16:48.5037
204.890 119.840 190.462770000 41.293643944 18 12:41:51.0648 +41:17:37.1182
640.430 740.800 190.425675000 41.243610388 9 12:41:42.1620 +41:14:36.9974
333.690 73.920 190.449105000 41.296123500 15 12:41:47.7852 +41:17:46.0446
At this stage I can make the wcs_setup.out file (like in example 1):
% cutup_xyrdnames hetacm_n4625.fits
Coordinate data file = hetacm_n4625.xyrd
% cat wcs_setup.out
10
565.160 497.970 190.430670000 41.263141638
13
433.350 455.450 190.443465000 41.266757972
8
665.790 410.090 190.419735000 41.268272389
7
704.050 255.610 190.414305000 41.280139917
18
204.890 119.840 190.462770000 41.293643944
9
640.430 740.800 190.425675000 41.243610388
15
333.690 73.920 190.449105000 41.296123500
--------------------------------------------------------------------
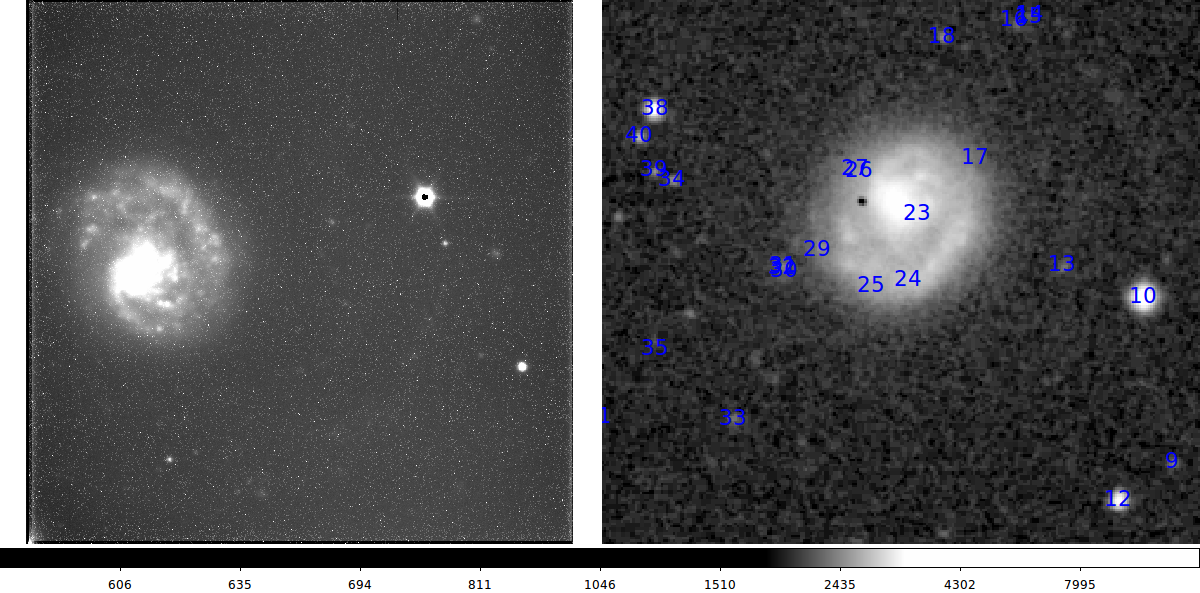
Below I show two finding chart from the above exercise.
 |
|
My initial usno_identify view of my NGC4625 acm image (left) and the
corresponding DSS view (right).
|
 |
|
My usno_identify view of my NGC4625 acm image (left) and the
corresponding DSS view (right). I have pulled a larger field DSS
image than the example above. The DSS identification labels changed because I
pulleds stars to a deeper magnitude limit.
|
First, since I have a good number of fit points, I run my well=tests
linear solution (wcs_2lin).
 |
Here is the result from my test wit wcs_2lin:
My wcs_setup.out file is present, so I run wcs_2lin:
% wcs_2lin hetacm_n4625.fits Y
My fit residuals in the final image displayed as:
Decision time, the final position residuals (in arcesconcds) are:
0.287 0.949 7 (RES_arcsec,Nstars)
|
The full wcs_trs treatment of the same data above did not work to completion.
I was creating a WCS that did not properly account for the image flip. The firast place to
start was to look at the trs_ solution first. Here is an example of how I convinced
myself that this part of the code was performing properly.
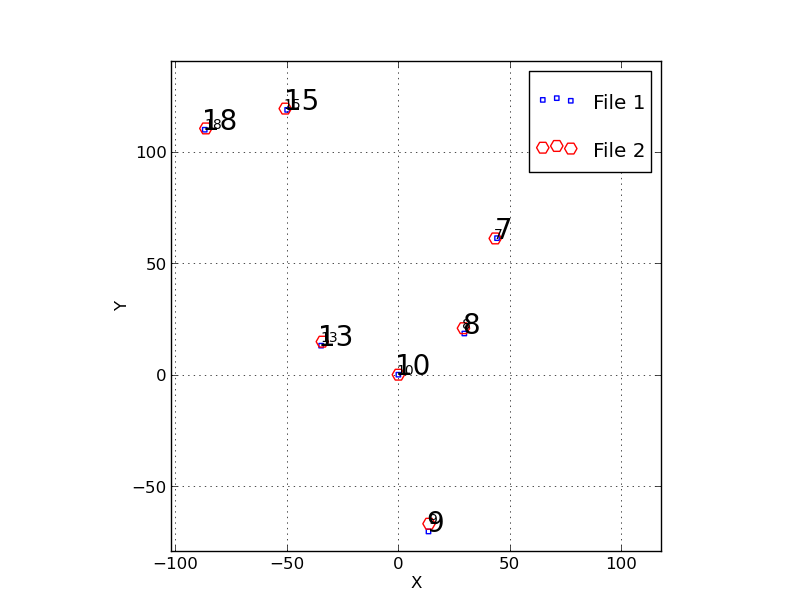 |
To diagnose a problem I was having with image produced initially by
wcs_trs I used my trs_ plotting package named trs_2plot. The code that
performs the fit is called trs_solve_2, and it produced a bunch of *.xy
files. These are explained in the file "Explain_Files.trs_solve_2". I
made a number of plots that showed me that, as far as trs_ was concerned, the
coordinate flip, scaling and rotation were being performed correctly. I made the
plot above that shows the original coordinates of the RA,DEC projected onto the
tangent plane in units of arcseconds (the small blur squares). The large red circles
are my image coordiantes in pixel units after conversion to the sky system using a
parity flip, a translation, a scale, and a rotation. Here is how I masde the plot:
% trs_2plot file1_trans.xy file2_rotate.xy
I made this in a subdirectory, so I had to rmember to copy the
PointNames file to there as well as the *.xy files.
The points line up quite well, which accounts for why my rms residuals
looked fine in the fit when I ran wcs_trs. Hence, it would seem that my
problem(s) are downstram: my routines that build and install the new
header are not working somewhere. By the way, I review in a laborous and
verbose way how to use trs_ plotting routines in my Big_Codes section
that discusses the whole trs_ methodology:
file:///home/sco/sco/scohtm/scocodes/trs_/trs_plotting.html#SEC_2
I don't link to the URL directly here since this doc will almost
certainly be moved around soon. But you get the idea.
|
Back to SCO CODES page




