Radial Surface Brightness Profiles
Here we discuss how to compute a radial profile. This can be used
to describe a stellar PSF or the light distribution in a galaxy.
Today the more frequently used approach is to fit the 2-D light
distribution in such source using an analytical model and some
optimization method. There is still a lot of utility in using
the 1-D approach, and floowwing the "walk before you run" approach,
I'll discuss some ways that I do that here.
- A very simple graphical approach to profiles.
- Some easy improvements.
A very simple graphical approach to profiles.
In computing a 1-D (one dimensional) surface brightness profile
for an image source, we are assuming the image has a symmetric radial
light distribution. By this I mean contours computed at different have
a circular of elliptical shape, and the center of each contour is
the same. This is not always the case , as we'll see in our
first exercice here with NGC3379, but is common enough for
well-behaved images of stars and poorly resolved galaxy images.
To begin, we'll look at some initial profile calculations I
made using the simple graphic tool named ds9_profiles. The
two main things we need for a profile is a center and an extent.
The ds9_profiles tool allows us to use a ds9 regions marker
to specify both on an image. We can also use markers to
sky sky boxes and maybe some other useful properties in the image.
Here are some notes from a "raw" run with ds9_profiles using a
single B-band image of NGC3379 (named Rsco2042.fits). I include
warts and all here to indicate some things might do to fix up the
code and make it a little easier to use.
Usage: ds9_profiles dsi11.fits 120.0 LOCAL_SKY N
Usage: ds9_profiles dsi11.fits 0.0 FIXED_SKY N
arg1 - name of input FITS image file
arg2 - asemi_max (= 0.0 to use individual region sizes)
arg3 - sky (= FIXED_SKY, fits_image_name, LOCAL_SKY)
arg4 - file cleanup (Y/N)
% ds9_profiles Rsco2042.fits 0 FIXED_SKY Y
Fails, but I save: p1.reg
I can load it and use it in future runs.
Use same regions, but:
% ds9_profiles Rsco2042.fits 0.0 FIXED_SKY Y
So, we must specify using the region ellipse semi-major axis
by saying R="0.0", not just R="0"
The actual peak pixel position for N3379 is X,Y = 1049,965
I fix this position, and a better ellipse, in: p2.reg
Yet, when I plot the growth curve, the title is
telling me different: X,Y=989,1037
Also, I get a shitty looking profile. What's up?
Now I just turn off the clean-up request (arg4) and I see
where that bogus center comes from:
% cat profpars.out_explain
# Output columns from profpars (using midodata.2)
Col01 = X center, pixels (from centroid)
Col02 = Y center, pixels (from centroid)
Col05 = semi-major axis length in pixels
Col06 = b/a axis ratio
Col07 = position angle (degrees) of major axis
Col08 = local background
Col08 = mean background (signal per pixel)
Col08 = mean error of background (signal per pixel)
% cat profpars.out
989.300 1037.690 137.412 0.87734 -64.277 230.8790 222.2015 6.1598
sky is okay
So, the local sky look reasonable, but the X,Y are not what I
thought I was setting via the regions file. What is going on?
The answer seems to be:
I display and set regions, but then I use profpars.sh to get the
ACTUAL profile parameters (which are always in a local file that
is named "profpars.out"). For my one ellipse here I get:
% cat profpars.out_explain
# Output columns from profpars (using midodata.2)
Col01 = X center, pixels (from centroid)
Col02 = Y center, pixels (from centroid)
Col05 = semi-major axis length in pixels
Col06 = b/a axis ratio
Col07 = position angle (degrees) of major axis
Col08 = local background
Col08 = mean background (signal per pixel)
Col08 = mean error of background (signal per pixel)
% cat profpars.out
989.300 1037.690 137.412 0.87734 -64.277 230.8790 222.2015 6.1598
**** And here is my probable answer: the centroid center,
which involves the WHOLE ellipse is not on the galaxy
peak (nucleus). What is the best (i.e. clearest to the
user) way to fix the profile center on the PEAK?
Some of the results from above are summarized in the two figure
immediately below.
 |
|
The ds9 view of Rsco2042.fits, our test image of NGC3379.
With ds9_profiles we have displayed the image and marked NGC3379
with an ellipse marker. We have also specified 4 sky boxes
that we'll use (via the MIDO routine inside ds9_profiles) to
compute a global mean background value for the image.
|
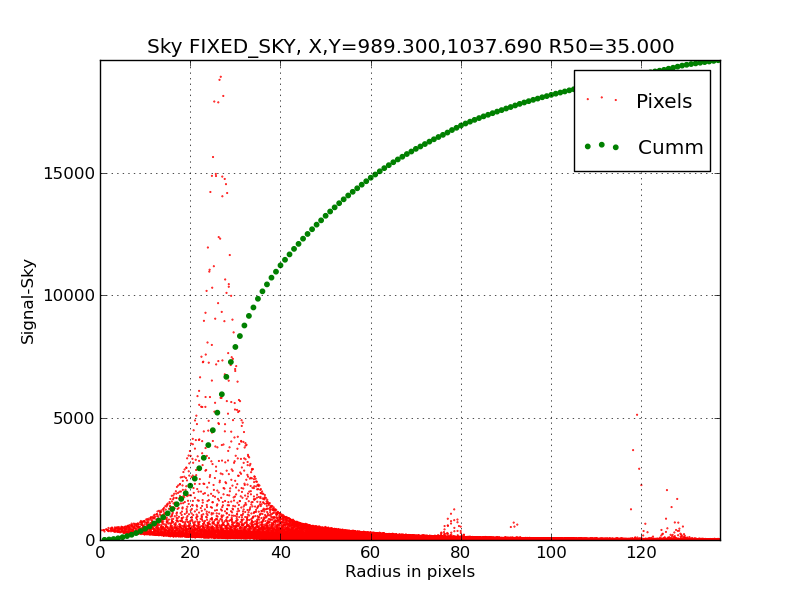 |
Here is the profile we derive from our first run of ds9_profiles
using the regions specified in the figure above. The command I
used was:
% ds9_profiles Rsco2042.fits 0.0 FIXED_SKY Y
We've used the global mean sky value for the image (FIXED_SKY) as
opposed to the local value around our ellipse regin (LOCAL_SKY). As
we see, the resulting profile is not very pleasing. The large "bump"
of excess light in the radius range 20pix to 40 pix is caused by
the fact that our centroid center (X,Y=989,1038) is really quite
far from the real position of the nuclues (X,Y = 1049,965). So, our
first question becomes: how can we compose a practical way of
"telling" ds9_profiles that we want to use the nucleus position
instead of the centroid center. A broader question might be: how often
does such a thing happen? The light distribution of NGC3379 is
extremely smooth and symmetric compared to most galaxies. Maybe this
sort of thing will be quite common. Notice alos the there are a
few more "bumps" of light in the outer extent of the profile. These
are bright stars in the image that have nothing to do with the
light from NGC3379. How should we eliminate the effect of these
stars? See below for the full explanation, but the
biggest error in making the above profile had to do with some bad header
cards and causing a misplaced radius model in the rad.fits image made
by the ellradius.sh code.
|
We can make a profile plot, but its use is limited. Before proceeding
with more a complicated analysis, possibly involving, contour fitting,
peak finding, and other things, maybe a few simple additions to
ds9_profiles would be worth considering:
- I could make the code recognize a default regions file
from the start? This would speed things up for the user,
and let them more easily build more and more complicated
region files.
- I can just give the user the option of hand editing
the profpars.out file. Just a low-tech for approach for
now.
Some easy improvements.
Based on the last considerations above, I have decided to quickly
review the guts of ds9_profile (as it stands now) and discuss some
easy ways to attain versatility and clarity during use. Here is what
happens when we execute ds9_profiles:
- The ccdsize script is used to derive the size of the input image.
- If a "midodata.2" file is NOT present, then we open a ds9 gui with
ds9_open (bash), display the input image with imlook (bash), set region
markers with ds9_id (bash), then run profpars.sh (OTW) to compile
paramters for regions we'll derive profiles for (currently
ellipse and circle markers).
- For each entry in the file "profpars.out" (from profpars.sh)
we build a radius map with ellradius.sh (OTW), collect the
profile points with profile_pnts.sh (OTW), and compute a growth
curve with profile_gcurve.sh (OTW).
- Lastly, for each profile, we construct a plot with
profile_plot_01.py (python) and display it interactively.
There are a lot of steps in #2 above, but it turns out that we can
make a couple simple changes to "imlook" that will help us out. First,
if we rename one of our region files (like p2.reg) to be
"Default.reg", I have altered imlook so that it will automatically
plot regions in that file. This gets us our "wish #1" at the
end of the last section. Moreover, in the process of adding this
to imlook, I added another similar feature. If the local file
"Default.greyscale" is present, the (z1,z2) values for
setting the image display greyscale are read from that
file used. The imlook tool attempts to derive a reasonable
scale, but sometimes we just wish to specify the (z1,z2) levels
for an image and leave it at that. I have added a discussion of
these features to my documentation
on imlook.
A common bonehead error!
As often happens, I had some "bad" header cards in my test image:
Rsco2042.fits. At least one of the cards was throwing off my coordinates, and
some of the bad features of my profile were due to my improper placement
of the radius image model. I used
the routine remove_fits_kwords to
fix this up. Briefly, here is what I did:
% cat bad.keys
CCDSEC
ORIGSEC
PROGRAM
DETECTOR
SSI
PREFLASH
GAIN
RDNOISE
CAMTEMP
DEWTEMP
CCDSUM
% remove_fits_kwords Rsco2042.fits bad.keys
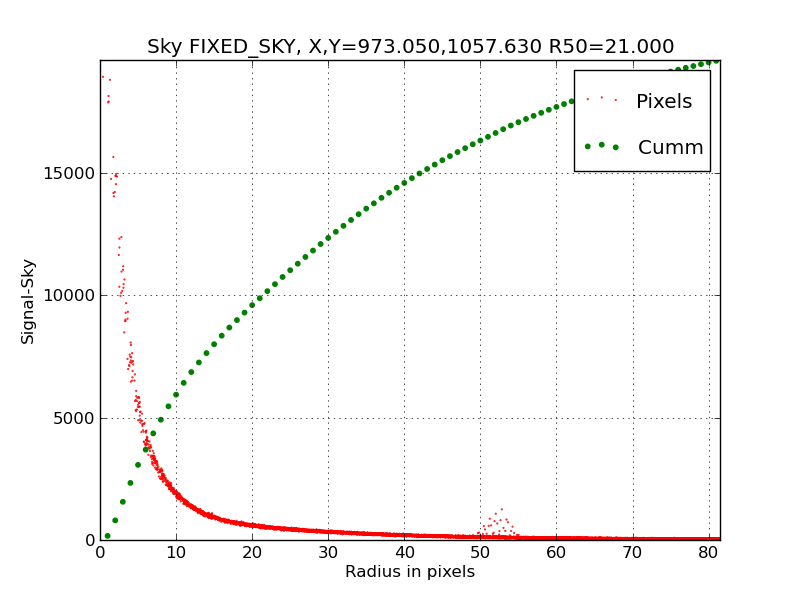
Having fixed my FITS header issues, I get a much more well-behaved
profile for NGC3379. Here is the figure, using a slightly smaller
semi-major axis size.
 |
|
Now this is more like it! Here we see the ds9_profiles result
after I have used the remove_fits_kwords routine to delete a bunch
of bullshit FITS header cards that were causing differences
between the image and physical coordinate systems. This was
causing the radius image model to be placed incorrectly. Hence,
my profile and growth curve estimates were completely messed up.
However, the changes I have added to ds9_profiles remain valid and
useful. Note that we would still see some "splitting" of the profile
points in the inner region we used the centroid center at
X,Y=974.1,1056.8 (see figure directly below). In this case, I altered
the ellipse region center based on the peak value and edited in
the nucleus position of X,Y=973.4,1057.2, and we see a much smoother
profile.
|
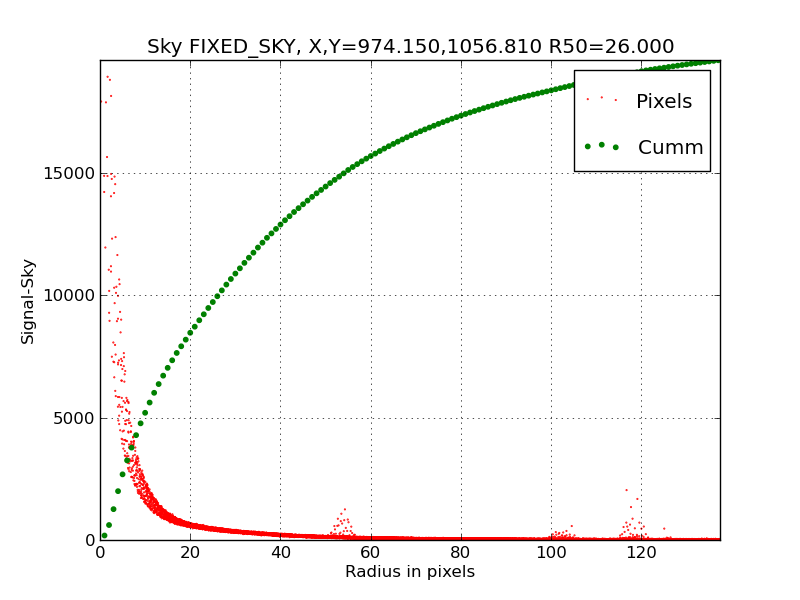 |
|
Here we have used the LOCAL_SKY and the ellipse region paramteres
defined by the large outer ellipse. The centroid center at
X,Y=974.1,1056.8 creates an "offset nucleus" situation in the inner
profile. A graphical means of assigning a "user-specified" center
is our next goal in our set of simple additions to ds9_profiles.
|
Finally, I have added the ability to graphically specify profile centers
using ds9_profiles. In the figure below I show and example of this.
For this set of regions I would create 3 profile plots. The red ellipse
markers serve as center specifiers for the larger green ellipse regions.
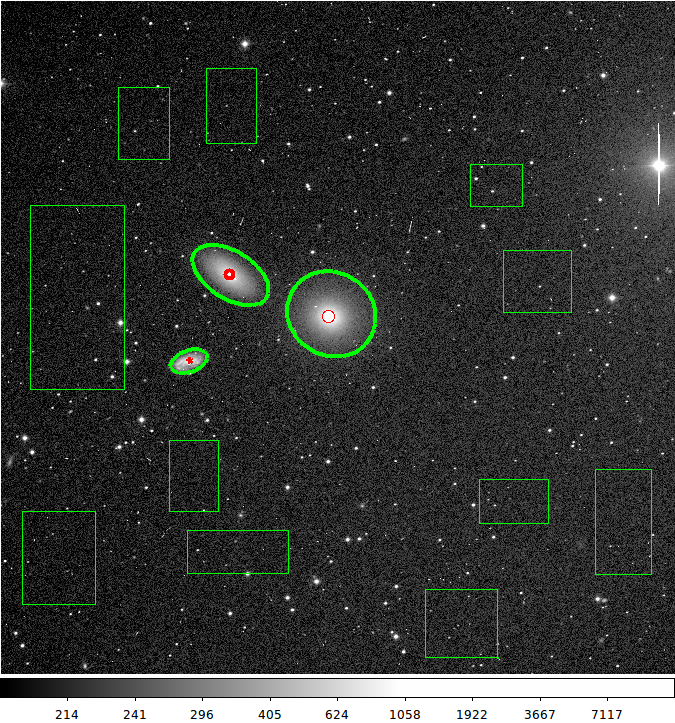 |
Here we have used the (new as of Nov2015) ability of
ds9_profiles to specify the center of an ellipse profile
aperture. The small (red) ellipse markers are used to isolate
the (X,Y) center we want for each elliptical profile, The colors
do not have to be red, but I made them that way here to
illustrate more clearly what is happening. This image and the
markers seen above can be reporduced withthe TDATA
exercise in:
$tdata/T_runs/profiles/ex1_n3379
|
Now we have a fairly simple way to compute radial profiles using
circle or ellipse regions.
Back to previous page




