A simple exercise after fits_review
I uses a simple exercise after a run of fits_review to make the figure below.
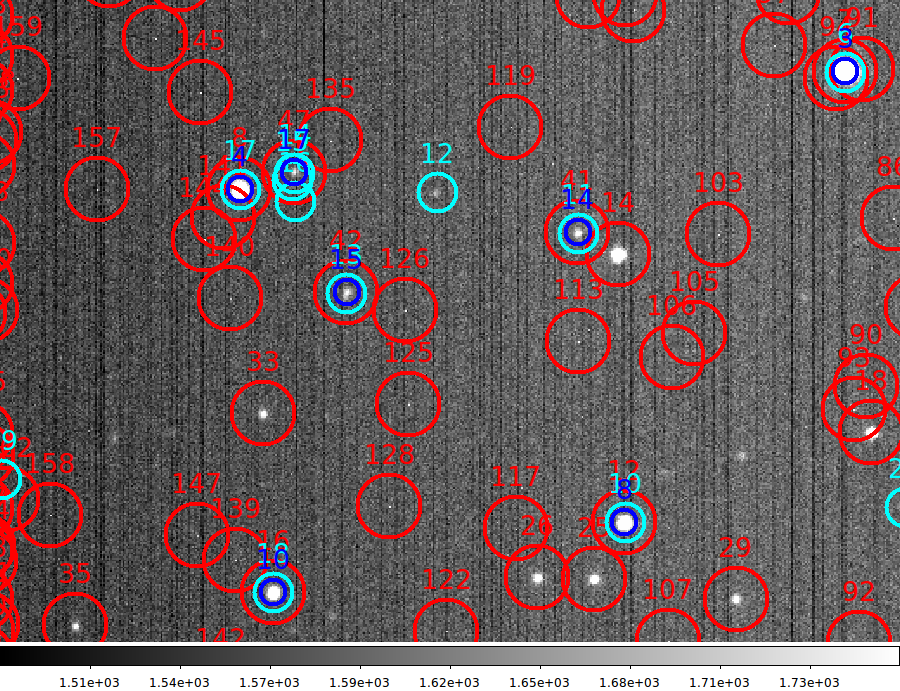 |
A view of an image we have fully processed with fits_review.
% cdfp2reg.sh 20170903T095516.8_acm_sci.cdfp 5.0 red n n
% cdfp2reg.sh 20170903T095516.8_acm_sci-usno.cdfp 3 cyan n n
% ds9_open 900 900
% ds9_view 20170903T095516.8_acm_sci.fits zscale n 1
I manually displayed the region files named:
20170903T095516.8_acm_sci-usno.cdfp.reg
20170903T095516.8_acm_sci.cdfp.reg
Since I had two cdfp files, I could easily cross-match the two sets by
Ra,Dec using the routine cdfp2reg.sh, which in this case ran almost
instantaneously. In the plot above, the RED circle indicate imgcat0
detections (5 sigma sky threshold) and the CYAN circles represnt sources
from the USNO-B1.0 catalog (not down to a faint limit). Finally, the
BLUE circles represent the targets that were positionally matched.Although
a match radius of 5 arcsec was used, the rms scatter of the match residuals
was in the range of a few thenths of an arcsec in both the Ra and Dec directions.
|
This file was originally: /home/sco/acm_play/20170903/red0/A5/S/README
The runs are made in /home/sco/acm_play/20170903/red0/A5
to get the data: ./S/GDAT
For a single acm image, here is what I have after a complete fits_review run:
20170903T095516.8_acm_sci.fits == image with a "rough" WCS installed
20170903T095516.8_acm_sci_detsig.fits
20170903T095516.8_acm_sci_bkg1.fits
20170903T095516.8_acm_sci_label0.fits
20170903T095516.8_acm_sci_sigma.fits
20170903T095516.8_acm_sci.cat0 == table file on imgcat0 run
20170903T095516.8_acm_sci.cdfp == a cdfp version of above (using WCS)
20170903T095516.8_acm_sci-usno.cdfp == the USNO catalog (cdfp)
I made region files from the *.cdfp and viewed them on the image using:
% cdfp2reg.sh 20170903T095516.8_acm_sci.cdfp 5.0 red n n
% cdfp2reg.sh 20170903T095516.8_acm_sci-usno.cdfp 3 cyan n n
% ds9_open 900 900
% ds9_view 20170903T095516.8_acm_sci.fits zscale n 1
The region files were named:
20170903T095516.8_acm_sci-usno.cdfp.reg
20170903T095516.8_acm_sci.cdfp.reg a cdfp version of above (using WCS)
To get cross-match and view the results:
% cdfpmatI.sh 20170903T095516.8_acm_sci.cdfp 20170903T095516.8_acm_sci-usno.cdfp 5.0
% cdfp2reg.sh cdfpmatI.cdfp 2 blue n n
Finally, I zoomed to an interesting part of the image and made: Fig1.png
In early Oct I seumed the development of the wcs_cdfp_refit code.
These notes and srcipts originally in: /home/sco/acm_play/20170903/red0/A6/S/README.A6
/home/sco/acm_play/20170903/red0/A6/S
- Use ./S/GRAB to get image and cdfp files
- make the positional crossmatch
cdfpmatI.cdfp 20170903T095516.8_acm_sci.cdfp 20170903T095516.8_acm_sci-usno.cdfp 5.0
- Make plots (xyplotter_prep, etc....)
xp cdfpmatI.cdfp a.table # edit
xyplotter_prep a.table 1
*** plot columns 11,12
*** Begin building the driver script based on input image name
- wcs_cdfp_refit blah.fits Y N
- Script BIG1, placed 2 levels up in the amin directory, will
run all images and overplot cross-matched catalogs, etc....
Oct12,2017
Resuming the development of wcs_cdfp_refit
% wcs_cdfp_refit blah.fits Y N
arg1 - Name of file with list of FITS images (must have cdfp files)
arg2 - Allow graphical review? (Y/N)
arg3 - Run in verbose/debug mode? (Y/N)
New code I am developing for rejection:
flag_outliers:
Flag lines in a tbale file based on a single user-specified column.
Usage: flag_outliers cdfpmatI dRA_arcsec 3.0 median 3
arg1 - Basename of table file (A1 for A1.table, must have params file)
arg2 - name of parameter
arg3 - beta rejection factor (3.0)
arg4 - mean or median
arg5 - number of rejection cycles
get_table_colnum:
Take as input the basename of a table file and the name of the
parameter you wish tolocate. If the baseneame is A.table, then a
corresponding A.params file must be present. The column number
of the params entry in the table is returned.
Usage: get_table_A1 basename Xcent
Usage: get_table_colnum A1 Xcent
arg1 = basename of the table file (e.g. A1 for A1.table)
arg2 = name of parameter (must be in existing A1.params file)
stats_outliers.sh:
Read values and compute statistical. Find outlier points
based on these statistics.
Usage: stats_outliers.sh X.dat mean 3.0 2
arg1 - name of input file with X values
arg2 - rejection about mean,median
arg3 - beta rejection level (beta-sigma)
arg4 - number of iterations
**** Bootom line: I can now address a lot of table file things using the
parameter name, not just the column number (but colget.py
still only takes the column number).
To combine the rejection flag files:
% combine_outlier_flags.sh list.c1 any
Usage: combine_oulier_flags.sh flag.list any
arg1 - name of input file listing flag files
arg2 - combine method (any,all)
To test this code use file in:
/home/sco/acm_play/20170903/red0/A6/p2
For example, I combine using "any" If any flag is set for a point, reject it
% combine_outlier_flags.sh list.c1 any
20 3 17 (Number_original,Number_flagged,Number_final)
The output file is: combine_outlier_flags.flags
It has 20 points, 3 of which are flagged.
Next I worked on using the combined flag file. I wanted to gathere a set
of X,Y,Ra,Dec for UNREJECTED targets. The form for this file, and hoe to
get it into my my wcs code was rather muddled. Below are my initial (and
muddled) notes on this.
wcs_markII = general wcs code , take xyrd file as input
How to make the xyrd file?
get_xyrd_from_reg == assemble the xyrd file (but where used?)
**** It looks like the wcs_markII can build the xyrd file
See the lines in wcs_markII:
#==================================================================
# Creat the basic input file = wcs_setup.out
# Recall that cutup_xyrdnames must be fed the name of the
# FITS file being handled (not the *.xyrd file name)
cutup_xyrdnames $basename.fits
#==================================================================
cutup_xyrdnames:
Take information from a cdfp_xyrdnames file and build
an [x,yr,d] file in the style of my older (pre-2016)
wcs codes. The name of this file was usually hard-coded
to be "wcs_setup.out". I reatin that name here.
To see how I create a cdfp_xyrdnames file, look at the
OTW code cdfp_xyrdnames.sh and scripts that call it like
ds9_midomark.
Another code to look at is the OTW code cdfp_xyrdnames.sh
----------------------------------------------------------------------
I think I see:
1) wcs_markII needs a wcs_setup.out file
2) I use cutup_xyrdnames to assembe wcs_setup.out from the
image name (image.xyrd is a file).
So, I need to understand (and better document) the cdfp_xyrdnames.sh code.
There is a HELP file, and it gives me the basic structur of the file I
need for cutup_xyrdnames:
The header gives the name of the input files, and explains the columns
of the output data table:
Col01 - X centroid from mido (physical units in the CCD image)
Col02 - Y centroid from mido (physical units in the CCD image)
Col03 - RA in units of degrees from USNO-B1 (echo $USNOBDIR)
Col04 - DEC in units of degrees from USNO-B1 (echo $USNOBDIR)
Col05 - Integrer name of source (line number in the cdfp file)
Col06 - RA (J2000) in sexigecimal format
Col07 - DEC (J2000) in sexigecimal format
The file that cutup_xyrdnames needed is the FITS name. The code gets the
image basename and then builds the name of the input file needed: basename.xyrd
This file is made with cdfp_xyrdnames.sh (which also has a HELP file).
SO: I need something for wcs_cdfp_refit that makes this *.xyrd file
Sun Oct 15 13:41:11 CDT 2017
============================
I have decided to build: cdfp_xyrdnames_2.sh
It will build a *.xyrd file for use by cutup_xyrdnames in wcs_markII.
BOTTOMLINE: I need to make a *.xyrd file, and it has form:
header for the *.xyrd is made in cdfp_xyrdnames.sh
The lines are written in cdfp_xyrdnames.f to a hard-coded
file named cdfp_xyrdnames.out with the following:
write(luno,2901) x,y,radeg,decdeg,line,rs1,ds1
2901 format(2(f12.3,1x),1x,2(f14.9,1x),2x,i5,2x,a11,1x,a11)
Some final points:
1) wcs_markII wants a *.xyrd file and I have two ways to make it:
a) Use cdfp_xyrdnames that combines a cdfp file with region files collected
gaphically with things like ds9_2f_markII.....
b) Use cdfp_xyrdnames_2 to collect points from a single cdfp (assembled with
cdfpmatI) but which skips over rejected points.
Wed Oct 18 10:27:03 CDT 2017
============================
Slogging on in: /home/sco/acm_play/20170903/red0/A6
% combine_outlier_flags.sh list.1 any $ to get final list of rejection flags
% mv combine_outlier_flags.flags Flags.final
To get the *.rdxy file:
% cdfp_xyrdnames_2.sh 20170903T095516.8_acm_sci.fits cdfpmatI.cdfp Flags.final
To compute the and install new wcs:
% wcs_markII 20170903T095516.8_acm_sci.fits 2lin
Back
